Psience.VPT2
An implementation of vibrational perturbation theory (VPT) that uses sparse matrix methods to obtain
corrections.
Makes heavy use of the BasisReps package as well as McUtils.Coordinerds to obtain representations
of the corrections to the vibrational Hamiltonian.
Is technically not restricted to VPT in a harmonic basis, but no other forms of PT are likely to
be implemented in the near future.
For the purposes of papers, we’ve been calling this implementation PyVibPTn.
The easiest way to run jobs is through the VPTRunner.run_simple interface.
The options for jobs along with short descriptions are detailed in
VPTSystem for molecule/system-related options,
VPTStateSpace for state-space & degeneracy-related options,
VPTHamiltonianOptions for expansion-related options,
VPTHamiltonianOptions for expansion-related options,
VPTSolverOptions for options related to constructing representations and applying VPT,
and VPTRuntimeOptions for options related to the runtime/logging
A basic tutorial to provide more extensive hand-holding can be found here.
Finally, the general code flow is detailed below
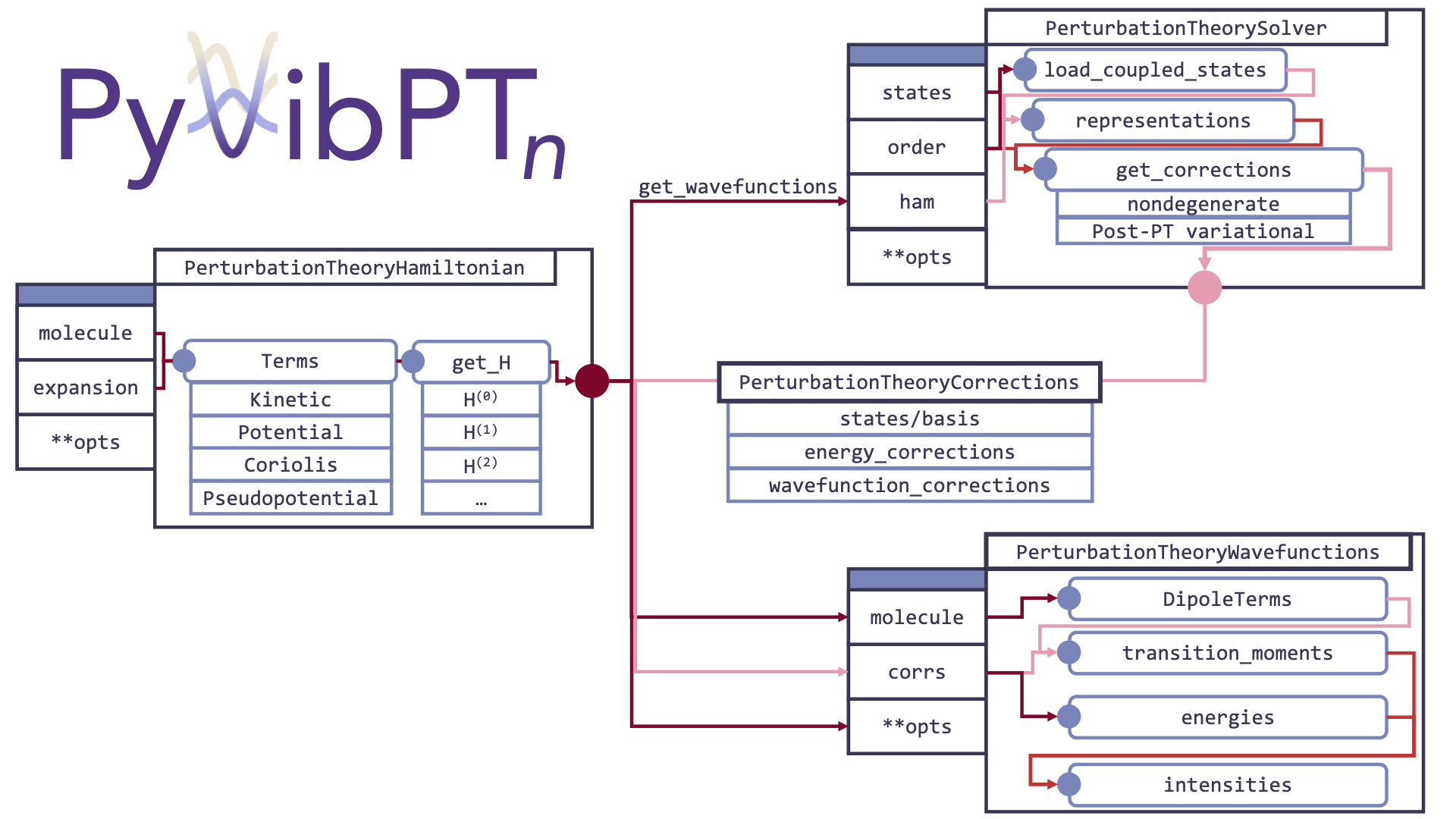
Members
The implementation of vibrational perturbation theory provided here uses a kernel/config/driver type of design.
The kernel that actually solves the perturbation theory equations is the PerturbationTheorySolver object.
The config comes the PerturbationTheoryHamiltonian, which implements the expansion of the Hamiltonian
with respect to normal modes.
Finally, the driver is the VPTRunner which through its run_simple method aggregates all of the possible options needed
for VPT and sends them to the right parts of the architecture.
Because of this, while it can be helpful to know how the PerturbationTheorySolver and PerturbationTheoryHamiltonian work, all that one
usually needs to do to run VPT is to call VPTRunner.run_simple.
The most basic call looks like
VPTRunner.run_simple(system_spec, states)
where the system_spec is often an fchk file from an electronic structure calculation that provides a restricted quartic potential
and the states is an int that specifies the max number of quanta of excitation in the states that will be corrected.
The system spec can also be more explicit, being either a Molecule object or a list like [atoms, coords, opts]
where atoms is a list of strings of the atoms, coords are the accompanying Cartesian coordinates, and opts is a dict
of options for the molecule, such as the masses.
It is also possible (and sometimes necessary) to supply custom normal modes, which can be done through the modes option (examples below).
Examples
In the following we provide some basic examples. More complex cases can be composed from the many settings provided in the Hamiltonian, solver, and runtime objects.
A note on input formats
A very common use case for this package is to extract more information from a VPT2 calculation
performed by some electronic structure package. To support this use case the Molecule
object can read in formatted checkpoint (.fchk) files and most of these examples will run off of those.
Running Jobs
The way to run jobs with the least boilerplate is to use the run_simple method of the VPTRunner class.
Run second-order vibrational perturbation theory for all states with up to three quanta of excitation and print their infrared spectroscopic data
VPTRunner.run_simple(
"HOH_freq.fchk",
3 # up through three quanta of excitation
)
...
Harmonic Anharmonic
State Frequency Intensity Frequency Intensity
0 0 1 3937.52466 67.02051 3744.74223 64.17167
0 1 0 3803.29960 4.14283 3621.97931 3.11401
1 0 0 1622.30302 67.45626 1572.70734 68.32367
0 0 2 7875.04932 0.00000 7391.41648 0.01483
0 2 0 7606.59919 0.00000 7155.85397 0.31496
2 0 0 3244.60604 0.00000 3117.39090 0.55473
0 1 1 7740.82426 0.00000 7200.36337 2.20979
1 0 1 5559.82768 0.00000 5294.37886 3.76254
1 1 0 5425.60262 0.00000 5174.61359 0.06232
0 0 3 11812.57398 0.00000 10940.02275 0.04985
0 3 0 11409.89879 0.00000 10601.62396 0.00898
3 0 0 4866.90906 0.00000 4634.05068 0.00350
0 1 2 11678.34892 0.00000 10680.67944 0.00001
1 0 2 9497.35234 0.00000 8917.98240 0.00333
0 2 1 11544.12385 0.00000 10567.87984 0.08362
2 0 1 7182.13070 0.00000 6815.99171 0.16303
1 2 0 9228.90221 0.00000 8688.41518 0.00427
2 1 0 7047.90564 0.00000 6699.22408 0.00661
1 1 1 9363.12728 0.00000 8729.92693 0.09713
It is can do the same by using the driver objects directly, but it is less convenient.
If one is interested in doing further analysis on the data, it is also possible to run jobs directly from a VPTAnalyzer to capture the output, like
analyzer = VPTAnalyzer.run_VPT("HOH_freq.fchk", 3)
The call structure is identical. The output is just routed through the analyzer.
The output tables can then be printed out using the analyzer’s print_output_tables method and various other properties can be requested.
For the complete set of properties, check the VPTAnalyzer documentation.
Internal Coordinates
It is possble to run the VPT in curvilinear internal coordinates. The most straightforward way to do this is to supply a Z-matrix (i.e. using polyspherical coordinates). For this, we say
VPTRunner.run_simple(
...,
internals=zmat
)
where zmat is an N-by-4 array of ints like
[
[atom1, bond1, angle1, dihed1],
[atom2, bond2, angle2, dihed2],
...
]
where the atom specifies which of the atoms from the original set of Cartesian coordinates we’re describing (0 for the first atom, 1 for the second, etc.), bond specifies which atom it is bonded to, angle provides the third atom for making an angle, and dihed provides the dihedral angle.
It is important to note that the first bond distance, the first two angles, and the first three dihedrals aren’t well defined and are generally used to store information about how the system is embedded in Cartesian space.
It is also possible to use functions of the internal coordinates. To do this we supply
internals={
'zmatrix': zmat,
'conversion': conv,
'inverse': inv
}
where conv looks like
def conv(r, t, f, **kwargs):
"""
Takes the bond lengths (`r`), angles `(t)` and dihedrals `(f)`,
and returns new coordinates that are functions of these coordinates
"""
... # convert the coordinates
return np.array([r, t, f])
and inv will take the output of conv and return the original Z-matrix/polyspherical coordinates.
- AnalyticPTOperators
- HOHVPTRunner
- HOHVPTSubstates
- BlockLabels
- ResultsFileAnalysis
- IHOHExcited
- HOHVPTAnneManip
- HOHPartialQuartic
- HOHVPTNonGSRunner
- HOHVPTRunnerFlow
- HOHVPTRunnerShifted
- OCHHVPTRunnerShifted
- HOONOVPTRunnerShifted
- CrieegeeVPTRunnerShifted
- HOHVPTRunner3rd
- GetDegenerateSpaces
- ClHOClRunner
- AnalyticModels
- HOHCorrectedDegeneracies
- HOHCorrectedPostfilters
- WaterSkippedCouplings
- H2COPolyads
- H2COModeSel
- HODRephase
- HOHRephase
- NH3
- HOONO
- H2COSkippedCouplings
- WaterDimerSkippedCouplings
- OCHHInternals
- NH3Units
- AnneAPI
Before we can run our examples we should get a bit of setup out of the way. Since these examples were harvested from the unit tests not all pieces will be necessary for all situations.
All tests are wrapped in a test class
class VPT2Tests(TestCase):
""" Threshold = 0
::> building ExpansionRepresentation<H(0)>
::> in Representation<T(0)>
> evaluating in BraKet space BraKetSpace(nstates=210)
> evaluating 210 elements over 21 unique indices sequentially
> took 0.127s
<::
::> in Representation<V(0)>
> evaluating in BraKet space BraKetSpace(nstates=210)
> evaluating 210 elements over 21 unique indices sequentially
> took 0.184s
<::
> took 0.520s
<::
::> building ExpansionRepresentation<H(1)>
::> in Representation<T(1)>
> evaluating in BraKet space BraKetSpace(nstates=1190)
> took 0.000s
<::
::> in Representation<V(1)>
> evaluating in BraKet space BraKetSpace(nstates=1190)
> evaluating 1190 elements over 56 unique indices sequentially
> took 0.204s
<::
> took 0.499s
<::
::> building ExpansionRepresentation<H(2)>
::> in Representation<T(2)>
> evaluating in BraKet space BraKetSpace(nstates=43)
> took 0.000s
<::
::> in Representation<V(2)>
> evaluating in BraKet space BraKetSpace(nstates=43)
> evaluating 43 elements over 126 unique indices sequentially
> took 0.657s
<::
::> in Representation<Coriolis(0)>
> evaluating in BraKet space BraKetSpace(nstates=43)
> evaluating 43 elements over 906 unique indices sequentially
> took 1.005s
<::
::> in Representation<V'(0)>
> evaluating in BraKet space BraKetSpace(nstates=43)
> evaluating identity tensor over 43 elements
> took 0.036s
<::
> took 2.244s
<::
<::
::> Energy Corrections
> State <0|dH(2)|0> <0|dH(1)|1>
0 0 0 0 0 0 -5.49879 -75.41416
0 0 0 0 0 1 48.22641 -294.08843
0 0 0 0 1 0 46.72377 -284.71337
0 0 0 1 0 0 2.02819 -114.86847
0 0 1 0 0 0 -32.88017 -81.93283
0 1 0 0 0 0 -34.90506 -66.80892
1 0 0 0 0 0 -46.81545 -55.50575
0 1 0 1 0 0 -25.19818 -114.53080
0 0 0 0 0 1 3061.70147 95.24194 2849.45769 63.44723
0 0 0 0 1 0 2977.64050 69.29750 2820.56385 64.99539
0 0 0 1 0 0 1727.08265 63.79277 1695.15532 65.08450
0 0 1 0 0 0 1527.04079 11.12160 1493.14075 9.28519
0 1 0 0 0 0 1252.16397 9.69252 1231.36294 10.18280
1 0 0 0 0 0 1188.11375 7.01998 1166.70551 7.08986
0 1 0 1 0 0 2979.24662 0.00000 2967.72530 43.44534
"""
""" Threshold = 0.05 cm^-1
0 0 0 0 0 1 3061.70147 95.24194 2849.45684 63.44730
0 0 0 0 1 0 2977.64050 69.29750 2820.56243 64.99418
0 0 0 1 0 0 1727.08265 63.79277 1695.15532 65.08644
0 0 1 0 0 0 1527.04080 11.12160 1493.14075 9.28423
0 1 0 0 0 0 1252.16397 9.69252 1231.36294 10.18282
1 0 0 0 0 0 1188.11376 7.01998 1166.70551 7.08986
0 1 0 1 0 0 2979.24662 0.00000 2967.72474 43.44434
::> building ExpansionRepresentation<H(0)>
::> in Representation<T(0)>
> evaluating in BraKet space BraKetSpace(nstates=144)
> evaluating 144 elements over 21 unique indices sequentially
> took 0.089s
<::
::> in Representation<V(0)>
> evaluating in BraKet space BraKetSpace(nstates=144)
> evaluating 144 elements over 21 unique indices sequentially
> took 0.176s
<::
> took 0.449s
<::
::> building ExpansionRepresentation<H(1)>
::> in Representation<T(1)>
> evaluating in BraKet space BraKetSpace(nstates=287)
> took 0.000s
<::
::> in Representation<V(1)>
> evaluating in BraKet space BraKetSpace(nstates=287)
> evaluating 287 elements over 56 unique indices sequentially
> took 0.238s
<::
> took 0.559s
<::
::> building ExpansionRepresentation<H(2)>
::> in Representation<T(2)>
> evaluating in BraKet space BraKetSpace(nstates=21)
> took 0.000s
<::
::> in Representation<V(2)>
> evaluating in BraKet space BraKetSpace(nstates=21)
> evaluating 21 elements over 126 unique indices sequentially
> took 0.415s
<::
::> in Representation<Coriolis(0)>
> evaluating in BraKet space BraKetSpace(nstates=21)
> evaluating 21 elements over 906 unique indices sequentially
> took 0.506s
<::
::> in Representation<V'(0)>
> evaluating in BraKet space BraKetSpace(nstates=21)
> evaluating identity tensor over 21 elements
> took 0.118s
<::
> took 1.760s
<::
"""
""" Threshold = 1.0 cm^-1
::> building ExpansionRepresentation<H(0)>
::> in Representation<T(0)>
> evaluating in BraKet space BraKetSpace(nstates=144)
> evaluating 144 elements over 21 unique indices sequentially
> took 0.063s
<::
::> in Representation<V(0)>
> evaluating in BraKet space BraKetSpace(nstates=144)
> evaluating 144 elements over 21 unique indices sequentially
> took 0.142s
<::
> took 0.582s
<::
::> building ExpansionRepresentation<H(1)>
::> in Representation<T(1)>
> evaluating in BraKet space BraKetSpace(nstates=287)
> took 0.000s
<::
::> in Representation<V(1)>
> evaluating in BraKet space BraKetSpace(nstates=287)
> evaluating 287 elements over 56 unique indices sequentially
> took 0.262s
<::
> took 0.901s
<::
::> building ExpansionRepresentation<H(2)>
::> in Representation<T(2)>
> evaluating in BraKet space BraKetSpace(nstates=19)
> took 0.000s
<::
::> in Representation<V(2)>
> evaluating in BraKet space BraKetSpace(nstates=19)
> evaluating 19 elements over 126 unique indices sequentially
> took 0.336s
<::
::> in Representation<Coriolis(0)>
> evaluating in BraKet space BraKetSpace(nstates=19)
> evaluating 19 elements over 906 unique indices sequentially
> took 0.601s
<::
::> in Representation<V'(0)>
> evaluating in BraKet space BraKetSpace(nstates=19)
> evaluating identity tensor over 19 elements
> took 0.064s
<::
> took 1.756s
<::
<::
0 0 0 0 0 0 -4.96621 -75.41416
0 0 0 0 0 1 48.17888 -294.08843
0 0 0 0 1 0 46.58555 -284.71337
0 0 0 1 0 0 1.52477 -114.86847
0 0 1 0 0 0 -33.06100 -81.93283
0 1 0 0 0 0 -34.75406 -66.80892
1 0 0 0 0 0 -47.74137 -55.50575
0 1 0 1 0 0 -26.31829 -114.53080
0 0 0 0 0 1 3061.70147 95.24194 2848.44632 62.90510
0 0 0 0 1 0 2977.64050 69.29750 2819.89305 64.85348
0 0 0 1 0 0 1727.08265 63.79277 1694.11932 65.38942
0 0 1 0 0 0 1527.04080 11.12160 1492.42734 9.04394
0 1 0 0 0 0 1252.16397 9.69252 1230.98136 10.06742
1 0 0 0 0 0 1188.11376 7.01998 1165.24700 7.08479
0 1 0 1 0 0 2979.24662 0.00000 2966.50387 43.86153
"""
AnalyticPTOperators
def test_AnalyticPTOperators(self):
internals = False
vpt2 = AnalyticPerturbationTheorySolver.from_order(2, internals=internals)
# print(
# vpt2.hamiltonian_expansion[2].get_poly_terms([])
# )
# raise Exception(...)
#
# H1PH1 = vpt2.energy_correction(2).expressions[1]
#
# H1 = vpt2.hamiltonian_expansion[1]
# H1H1 = H1*H1
"""
==================== V[1](0, 0, 0)V[1](0, 0, 0) 1 ====================
:: 1
> 1 [array([0. , 0. , 0. , 1.125])]
> 1.1249999999999993 [array([1., 3., 3., 1.])]
> 0.7499999999999996 [array([1. , 1.83333333, 1. , 0.16666667])]
> 1 [array([ 0. , 0.25 , -0.375, 0.125])]
"""
# H1H1.get_poly_terms([],
# # allowed_paths=[
# # ((1,), (-1,)),
# # ((3,), (-3,))
# # ]
# ).prune_operators([(1, 1)]).print_tree()
# raise Exception(...)
PH1 = vpt2.wavefunction_correction(1).expressions[0]
E2 = vpt2.energy_correction(2)
# # pt_shit = H1PH1.get_poly_terms((4,)).combine()
# pt_shit = PH1((1,), simplify=False).expr.combine()
# raise Exception(
# PH1.get_poly_terms((1,)).terms[((1,0, 0, 0, 0),)].terms[((1,),)].coeffs
# )
# pt_shit = E2.expressions[1]([], simplify=False).expr
# raise Exception(pt_shit)
# subpoly = pt_shit.terms[((1, 1, 0, 2, 1), (1, 1, 1, 0, 2))].combine(combine_energies=False).terms[((1, 1, 1),)]
# for p in subpoly.polys:
# print(p.prefactor, p.coeffs)
# raise Exception(...)
# h3_poly = vpt2.hamiltonian_expansion[1]([3]).expr.terms[((1, 0, 0, 0, 0),)]
# raise Exception(
# h3_poly.prefactor,
# h3_poly.coeffs
# )
# with np.printoptions(linewidth=1e8):
# pt_shit = H1H1([]).expr
# pt_shit.print_tree()
#
# raise Exception(...)
# # raise Exception(
# # Y1.get_poly_terms((1,))
# # # [
# # # [p.prefactor for p in psum.polys]
# # # for psum in h1y1.get_poly_terms(()).terms.values()
# # # ]
# # )
#
# h1y1 = E2.expressions[1]
# # raise Exception(h1y1, h1y1.gen2, h1y1.gen2.expressions)
#
# raise Exception(
# h1y1.get_poly_terms(()).combine()
# # [
# # [p.prefactor for p in psum.polys]
# # for psum in h1y1.get_poly_terms(()).terms.values()
# # ]
# )
# for _ in vpt2.energy_correction(2).expressions[1].changes[()]:
# print(_)
# test_sum = list(jesus_fuck.terms.values())[1]
# # for poly in test_sum.polys:
# # print(poly.coeffs)
# raise Exception(test_sum, test_sum.combine())
# raise Exception(test_sum.polys[0].combine(test_sum.polys[1]).coeffs)
# load H20 parameters...
file_name = "HOH_freq.fchk"
from Psience.BasisReps import HarmonicOscillatorMatrixGenerator
HarmonicOscillatorMatrixGenerator.default_evaluator_mode = 'rho'
runner, _ = VPTRunner.construct(
TestManager.test_data(file_name),
1,
internals=(
[[0, -1, -1, -1], [1, 0, -1, -1], [2, 0, 1, -1]]
if internals else
None
),
logger=False,
zero_element_warning=False
# mode_selection=[0, 1]
)
ham = runner.hamiltonian
V = ham.V_terms
G = ham.G_terms
U = ham.pseudopotential_term
g2 = G[2]
if internals:
water_expansion = [
[V[0]/2, G[0]/2],
[
# np.zeros(V[1].shape),
np.sum([V[1].transpose(p) for p in itertools.permutations([0, 1, 2])], axis=0)/np.math.factorial(3)/6,
# np.zeros(V[1].shape),
-np.moveaxis(G[1], -1, 0)/2
if isinstance(G[1], np.ndarray) else
np.zeros(V[1].shape)
],
[
# np.zeros(V[2].shape),
V[2]/24,
# np.zeros(V[2].shape),
-np.moveaxis(G[2], -1, 0)/4
if isinstance(G[2], np.ndarray) else
np.zeros(V[2].shape),
# 0
U[0]/8
]
]
else:
Z = ham.coriolis_terms
water_expansion = [
[V[0] / 2, G[0] / 2],
[
np.zeros(V[1].shape),
# np.sum(
# [V[1].transpose(p) for p in itertools.permutations([0, 1, 2])],
# axis=0
# ) / np.math.factorial(3) / 6,
0 # G
],
[
# np.zeros(V[2].shape),
V[2] / 24,
0, # G
0, # V'
# np.zeros(V[2].shape),
-Z[0],
# 0
U[0] / 8 # Watson
]
]
# raise Exception(
# -.25 * np.array([
# Z[0][0, 0, 2, 2],
# Z[0][0, 2, 0, 2],
# Z[0][0, 2, 2, 0],
# Z[0][2, 0, 0, 2],
# Z[0][2, 0, 2, 0],
# Z[0][2, 2, 0, 0]
# ]) * 219475
# )
# raise Exception(
# -.25*np.array([
# Z[0][0, 0, 2, 2],
# Z[0][0, 2, 0, 2],
# Z[0][0, 2, 2, 0],
# Z[0][2, 0, 0, 2],
# Z[0][2, 0, 2, 0],
# Z[0][2, 2, 0, 0]
# ]) * 219475
# )
water_freqs = ham.modes.freqs
# raise Exception(list(sorted([
# [p, np.linalg.norm((np.transpose(water_expansion[2][3], p) - water_expansion[2][3]).flatten())]
# for p in itertools.permutations([0, 1, 2, 3])
# ],
# key=lambda x:x[1]
# )))
# solver = runner.hamiltonian.get_solver(runner.states.state_list)
# raise Exception(
# solver.representations[1].todense()[0][:4],
# solver.flat_total_space.excitations[:4]
# )
# wfns = runner.get_wavefunctions()
# ham.G_terms = [
# G[0],
# # G[1],
# np.zeros(G[1].shape),
# # G[2]
# np.zeros(G[2].shape),
# ]
# ham.V_terms = [
# V[0],
# # np.sum([V[1].transpose(p) for p in itertools.permutations([0, 1, 2])], axis=0) / np.math.factorial(3),
# np.zeros(V[1].shape),
# V[2],
# # np.zeros(V[2].shape)
# ]
# ham.coriolis_terms = [
# np.zeros(V[2].shape)
# # (Z[0] + np.transpose(Z[0], [0, 3, 2, 1])) / 2
# ]
# ham.pseudopotential_term = [0]
runner.print_tables(print_intensities=False)
"""
:: State <0|dH(2)|0> <0|dH(1)|1>
0 0 0 0.00000 -141.74965
0 0 1 0.00000 -514.12544
0 1 0 0.00000 -509.59810
1 0 0 0.00000 -165.17359
:: State <0|dH(2)|0> <0|dH(1)|1>
0 0 0 81.09533 -156.70145
0 0 1 276.68965 -545.08549
0 1 0 281.61581 -538.53511
1 0 0 88.49014 -213.67502
:: State <0|dH(2)|0> <0|dH(1)|1> # Cartesians
0 0 0 42.02414 -117.62974
0 0 1 199.53514 -467.93034
0 1 0 195.87334 -452.78667
1 0 0 -32.13943 -93.04841
>>--------------------------------------------------<<
>>------------------------- States Energies -------------------------
:: State Harmonic Anharmonic Harmonic Anharmonic
ZPE ZPE Frequency Frequency
0 0 0 4681.56362 4605.95750 - -
0 0 1 - - 3937.52464 3744.73491
0 1 0 - - 3803.29958 3621.98639
1 0 0 - - 1622.30304 1572.72428
>>--------------------------------------------------<<
"""
# V1 = water_expansion[1][0]
# G1 = water_expansion[1][1]
# print(G[1]/2)
# print(G1)
# raise Exception(...)
# raise Exception(
# wfns.corrs.wfn_corrections[1].todense()[0][:4],
# wfns.corrs.total_basis.excitations[:4],
# -7.14635718e-03 * water_freqs[0],
# -5.83925920e-03 * water_freqs[0],
# 0.02061049 * water_freqs[0],
# (
# V1[0, 0, 0]*1.060660171779821
# +(V1[0, 1, 1] + V1[0, 2, 2])*1.060660171779821
# + G1[0, 0, 0]*-0.35355339
# + (G1[1, 0, 1] + G1[2, 0, 2])*-0.35355339
# ) / water_freqs[0]
# # # wfns.corrs.wfn_corrections[1].todense()[0, 1],
# # # should be .005839259198189573
# # V[1][0, 0, 0]/water_freqs[0]*1.06066017
# # # + G[1][0, 0, 0]/water_freqs[0]*-0.35355339
# # + (V[1][0, 1, 1] + V[1][0, 2, 2])/water_freqs[0]*0.70710678*.5
# # # + (G[1][0, 1, 1] + G[1][0, 2, 2])/water_freqs[0]*0.70710678*-.5
# )
# 0.02819074222400865, -0.035337103975354986, [-0.00758025182870669, -0.027756852139690837]
# wfns = runner.get_wavefunctions()
# Y1 = vpt2.wavefunction_correction(1)
# raise Exception(
# wfns.corrs.wfn_corrections[1].todense()[0][:10],
# wfns.corrs.total_basis.excitations[:10],
# Y1([3,]).evaluate([0, 0, 0], water_expansion, water_freqs),
# Y1([2, 1]).evaluate([0, 0, 0], water_expansion, water_freqs),
# )
with np.printoptions(linewidth=1e8):
jesus_fuck = E2([])
# jesus_fuck.expr.print_tree()
corr = jesus_fuck.evaluate([0, 0, 0], water_expansion, water_freqs, verbose=True) * UnitsData.convert("Hartrees", "Wavenumbers")
print(corr)
raise Exception(corr)
# vpt2.energy_correction(2).expressions[1].changes[()]
raise Exception(
AnalyticPerturbationTheoryDriver.from_order(2).energy_correction_driver(
2
).get_poly_evaluator(
[ # tensor coeffs
[
np.eye(3),
np.eye(3),
],
[
np.ones((3, 3, 3)),
np.ones((3, 3, 3))
],
[
np.ones((3, 3, 3, 3)),
np.ones((3, 3, 3, 3)),
1
]
],
[1, 1, 1] # freqs
)
)
corrections = AnalyticPTCorrectionGenerator([
[
['x', 'x', 'x'],
['p', 'x', 'p']
],
# ['x'],
[
['x', 'x', 'x'],
['p', 'x', 'p']
]
]).get_correction([2])
v1 = np.ones((3, 3, 3))
g1 = np.ones((3, 3, 3))
return corrections.evaluate([0, 0, 0], [[v1, g1], [v1, g1]], [1, 1, 1], 1)
raise Exception(corrections)
coeffs = np.array([
TensorCoeffPoly({((1, 0, 0),):2, ((0, 1, 0),):1}),
TensorCoeffPoly({((0, 0, 1),):1}),
], dtype=object)
# new_b_poly = np.dot(
# [[1, 3], [2, 1]],
# coeffs
# )
# raise Exception(
# np.dot(
# [[-1, 3], [2, -1]],
# np.dot(
# [[1, 3], [2, 1]],
# coeffs
# )
# )/5
# )
# from Psience.AnalyticModels import AnalyticModel
#
# AnalyticModel(
# [
# AnalyticModel.r(0, 1),
#
# ]
# ).g()
from McUtils.Zachary import DensePolynomial
shifted = DensePolynomial([
[1, 2, 3, 4, 5],
[1, -2, 0, 6, 8]
]).shift([2, 3])
# raise Exception(shifted.deriv(1).coeffs)
self.assertTrue(
np.allclose(
shifted.coeffs,
[
[2157., 2716., 1281., 268., 21.],
[ 805., 1024., 486., 102., 8.]
]
)
)
#
new = DensePolynomial(coeffs)*DensePolynomial(coeffs)
raise Exception(new)
# """
# DensePolynomial([TensorCoeffPoly({((1, 0, 0), (1, 0, 0)): 4, ((0, 1, 0), (1, 0, 0)): 4, ((0, 1, 0), (0, 1, 0)): 1},1)
# TensorCoeffPoly({((0, 0, 1), (0, 1, 0)): 2, ((0, 0, 1), (1, 0, 0)): 4},1)
# TensorCoeffPoly({((0, 0, 1), (0, 0, 1)): 1},1)], 1)"""
# raise Exception(
# PTPoly(coeffs)*
# PTPoly(coeffs)
# )
base_classes = RaisingLoweringClasses(14, [2, 4, 2])
print(list(base_classes))
HOHVPTRunner
def test_HOHVPTRunner(self):
file_name = "HOH_freq.fchk"
from Psience.BasisReps import HarmonicOscillatorMatrixGenerator
HarmonicOscillatorMatrixGenerator.default_evaluator_mode = 'rho'
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
memory_constrained=True,
logger=True
)
HarmonicOscillatorMatrixGenerator.default_evaluator_mode = 'poly'
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
memory_constrained=True,
logger=True
)
HOHVPTSubstates
def test_HOHVPTSubstates(self):
file_name = "HOH_freq.fchk"
mol = Molecule.from_file(TestManager.test_data(file_name),
internals=[[0, -1, -1, -1], [1, 0, -1, -1], [2, 0, 1, -1]])
# raise Exception(mol.internal_coordinates)
ics = mol.internal_coordinates
derivs = mol.coords.jacobian(
ics.system,
[1, 2],
all_numerical=True,
converter_options={'reembed': True}
)
derivs = [x.reshape((9,) * (i + 1) + (3, 3)) for i, x in enumerate(derivs)]
VPTRunner.run_simple(
# TestManager.test_data(file_name),
mol,
# 1,
[[0, 0, 0], [1, 0, 0]],
# [[0, 0, 0], [0, 2, 1]],
# 2,
order=2,
expansion_order=2,
# 3,
# expansion_order={'default':1, 'dipole':2},
# target_property='wavefunctions',
# internals=mol.zmatrix,
# initial_states=1,
# operators={
# 'OH1': [ics[1, 0], derivs[0][:, 1, 0], derivs[1][:, :, 1, 0]],
# 'OH2': [ics[2, 0], derivs[0][:, 2, 0], derivs[1][:, :, 2, 0]],
# 'HOH': [ics[2, 1], derivs[0][:, 2, 1], derivs[1][:, :, 2, 1]]
# },
logger=True
)
BlockLabels
def test_BlockLabels(self):
VPTRunner.run_simple(
TestManager.test_data("i_hoh_opt.fchk"),
VPTStateSpace.get_state_list_from_quanta(4, 6) + [
[0, 1, 2, 2, 0, 0]
],
initial_states=[
[0, 0, 0, 0, 0, 0],
[0, 0, 0, 2, 0, 0],
[0, 1, 0, 2, 0, 0],
[0, 0, 0, 0, 1, 0]
],
# degeneracy_specs='auto',
degeneracy_specs={
'wfc_threshold':.3
# "polyads": [
# [
# [0, 0, 0, 0, 1, 0],
# [0, 0, 0, 2, 0, 0]
# ],
# [
# [0, 0, 0, 1, 0, 0],
# [0, 0, 2, 0, 0, 0]
# ]
# ]
},
# target_property='wavefunctions',
logger=True,
# logger=os.path.expanduser("~/Desktop/specks/run.txt"),
plot_spectrum=False
)
ResultsFileAnalysis
def test_ResultsFileAnalysis(self):
temp_file = os.path.expanduser('~/Desktop/test_results.hdf5')
log_file = os.path.expanduser('~/Desktop/test_results.txt')
# os.remove(temp_file)
# wfns = VPTRunner.run_simple(
# TestManager.test_data("i_hoh_opt.fchk"),
# 2,
# plot_spectrum=False
# # initial_states=[
# # [0, 0, 0, 0, 0, 0],
# # [0, 0, 0, 2, 0, 0],
# # [0, 1, 0, 2, 0, 0],
# # [0, 0, 0, 0, 1, 0]
# # ]
# )
if not os.path.exists(temp_file):
VPTRunner.run_simple(
TestManager.test_data("i_hoh_opt.fchk"),
2,
# initial_states=[
# [0, 0, 0, 0, 0, 0],
# [0, 0, 0, 2, 0, 0],
# [0, 1, 0, 2, 0, 0],
# [0, 0, 0, 0, 1, 0]
# ],
# degeneracy_specs='auto',
degeneracy_specs={
"polyads": [
[
[0, 0, 0, 0, 1, 0],
[0, 0, 0, 2, 0, 0]
],
[
[0, 0, 0, 1, 0, 0],
[0, 0, 2, 0, 0, 0]
]
],
"extra_groups": [
[
[0, 0, 0, 0, 1, 0],
[0, 1, 0, 0, 1, 0],
[1, 0, 0, 0, 1, 0],
[0, 0, 0, 2, 0, 0],
[0, 1, 0, 2, 0, 0],
[1, 0, 0, 2, 0, 0],
[0, 0, 2, 1, 0, 0],
[0, 1, 2, 1, 0, 0],
[1, 0, 2, 1, 0, 0],
[0, 0, 4, 0, 0, 0],
[0, 1, 4, 0, 0, 0],
[1, 0, 4, 0, 0, 0]
]
]
},
# target_property='wavefunctions',
# logger=os.path.expanduser("~/Desktop/specks/run_wfns.txt"),
results=temp_file,
logger=log_file,
plot_spectrum=False
)
analyzer = VPTAnalyzer(temp_file)
# target_state = [0, 0, 2, 0, 0, 0]
# for i,block in analyzer.degenerate_states:
# # intersect([target_state], block) -> check non-empty
# ...
# McUtils.Numputils.intersection()
shifted_spec = analyzer.shifted_transformed_spectrum(
analyzer.degenerate_states[4], # 2 states, bend and OOP overtone
analyzer.deperturbed_hamiltonians[4], # 2x2 matrix
[0, -50 / UnitsData.hartrees_to_wavenumbers] # the shifts I want to add onto the diagonal
)
shifted_spec.plot()#.show()
print(shifted_spec.frequencies, shifted_spec.intensities)
with analyzer.log_parser as parser:
for i, block in enumerate(parser.get_blocks()):
for subblock in block.lines:
print(subblock.tag)
from McUtils.Scaffolding import LogParser
with LogParser(log_file) as parser:
for i, block in enumerate(parser.get_blocks()):
for subblock in block.lines:
print(subblock.tag)
IHOHExcited
def test_IHOHExcited(self):
wfns = VPTRunner.run_simple(
TestManager.test_data("i_hoh_opt.fchk"),
VPTStateSpace.get_state_list_from_quanta(4, 6) + [
[0, 1, 2, 2, 0, 0]
],
initial_states=[
[0, 0, 0, 0, 0, 0],
[0, 0, 0, 2, 0, 0],
[0, 1, 0, 2, 0, 0],
[0, 0, 0, 0, 1, 0]
],
# degeneracy_specs='auto',
degeneracy_specs = {
"polyads":[
[
[0, 0, 0, 0, 1, 0],
[0, 0, 0, 2, 0, 0]
],
[
[0, 0, 0, 1, 0, 0],
[0, 0, 2, 0, 0, 0]
]
],
"extra_groups": [
[
[0, 0, 0, 0, 1, 0],
[0, 1, 0, 0, 1, 0],
[1, 0, 0, 0, 1, 0],
[0, 0, 0, 2, 0, 0],
[0, 1, 0, 2, 0, 0],
[1, 0, 0, 2, 0, 0],
[0, 0, 2, 1, 0, 0],
[0, 1, 2, 1, 0, 0],
[1, 0, 2, 1, 0, 0],
[0, 0, 4, 0, 0, 0],
[0, 1, 4, 0, 0, 0],
[1, 0, 4, 0, 0, 0]
]
# [
# [0, 0, 0, 1, 1, 0],
# [0, 1, 0, 1, 1, 0],
# [1, 0, 0, 1, 1, 0],
# [0, 0, 0, 3, 0, 0],
# [0, 1, 0, 3, 0, 0],
# [1, 0, 0, 3, 0, 0],
# [0, 0, 2, 2, 0, 0],
# [0, 1, 2, 2, 0, 0],
# [1, 0, 2, 2, 0, 0],
# [0, 0, 4, 1, 0, 0],
# [0, 1, 4, 1, 0, 0],
# [1, 0, 4, 1, 0, 0]
# ]
]
},
target_property='wavefunctions',
logger=os.path.expanduser("~/Desktop/specks/run_wfns.txt"),
# logger=os.path.expanduser("~/Desktop/specks/run.txt"),
plot_spectrum=False
)
# raise Exception(wfns.initial_states, wfns.initial_state_indices)
multispec = wfns.get_spectrum().frequency_filter(600, 4400)
multispec.plot().savefig(
os.path.expanduser(f"~/Desktop/specks/full.pdf"),
transparent=True
)
for state,spec in zip(wfns.initial_states, multispec):
s = "".join(str(s) for s in state)
spec.plot(plot_range=[[600, 4400], [0, 500]], padding=[[0, 0], [0, 0]],
image_size=[.75 * 20, 5.48 * 20]
).savefig(
os.path.expanduser(f"~/Desktop/specks/state_{s}.pdf"),
transparent=True
)
multispec = wfns.get_deperturbed_spectrum().frequency_filter(600, 4400)
for state,spec in zip(wfns.initial_states, multispec):
s = "".join(str(s) for s in state)
spec.plot(plot_range=[[600, 4400], [0, 500]], padding=[[0, 0], [0, 0]],
image_size=[.75 * 20, 5.48 * 20]
).savefig(
os.path.expanduser(f"~/Desktop/specks/state_depert_{s}.pdf"),
transparent=True
)
HOHVPTAnneManip
def test_HOHVPTAnneManip(self):
runner, _ = VPTRunner.helpers.run_anne_job(
TestManager.test_data("vpt2_helpers_api/hod/r"),
return_runner=True,
order=2,
expansion_order=2
)
# runner, opts = VPTRunner.construct('HOH', 3)
# Collect expansion data from runner
H = runner.hamiltonian
V = H.V_terms
freqs = np.diag(V[0]) # how they actually get fed into the code...
G = H.G_terms
U = H.pseudopotential_term
D = H.expansion_options['dipole_terms'] # these usually get fed forward to the wave functions
# raise Exception([[x.shape if isinstance(x, np.ndarray) else x for x in D[a]] for a in range(3)])
# Define new shifted frequencies
frequency_shift = np.array([-1, 0, 0]) * UnitsData.convert("Wavenumbers", "Hartrees")
new_freqs = freqs + frequency_shift
# Rescale parameters
scaling_factor = np.sqrt(new_freqs) / np.sqrt(freqs)
# we use NumPy broadcasting tricks to rescale everything
G[2] # this requires the highest-order derivatives, so by doing it first and letting everything
# cache less junk gets printed to screen
v_expansion = [
# d V /dq_i dq_j
V[0] * (scaling_factor[:, np.newaxis] * scaling_factor[np.newaxis, :]),
# d V /dq_i dq_j dq_k
V[1] * (
scaling_factor[:, np.newaxis, np.newaxis] *
scaling_factor[np.newaxis, :, np.newaxis] *
scaling_factor[np.newaxis, np.newaxis, :]
),
# d V /dq_i dq_j dq_k dq_l
V[2] * (
scaling_factor[:, np.newaxis, np.newaxis, np.newaxis] *
scaling_factor[np.newaxis, :, np.newaxis, np.newaxis] *
scaling_factor[np.newaxis, np.newaxis, :, np.newaxis] *
scaling_factor[np.newaxis, np.newaxis, np.newaxis, :]
),
]
# For the momentum axes we _divide_ by the scaling factor
g_expansion = [
# Formally we should be dividing by this scaling factor, but to make sure
# V[0] == G[0] we multiply
# G_i,j
G[0] * (scaling_factor[:, np.newaxis] * scaling_factor[np.newaxis, :]),
# For the derivatives, we do the scaling correct
# d G_j,k / dq_i (i.e. q-index corresponds to axis 0)
G[1] * (
scaling_factor[:, np.newaxis, np.newaxis] /
scaling_factor[np.newaxis, :, np.newaxis] /
scaling_factor[np.newaxis, np.newaxis, :]
),
# d G_k,l / dq_i dq_j (i.e. q-indices correspond to axes 0,1)
G[2] * (
scaling_factor[:, np.newaxis, np.newaxis, np.newaxis] * # Note that we multiply for the first two axes
scaling_factor[np.newaxis, :, np.newaxis, np.newaxis] /
scaling_factor[np.newaxis, np.newaxis, :, np.newaxis] /
scaling_factor[np.newaxis, np.newaxis, np.newaxis, :]
)
]
# I haven't done the math to figure out how exactly u should transform
u_expansion = [U[0]]
d_expansion = [
[
D[a][0], # mu_a
# d mu_a / dq_i
D[a][1] * scaling_factor[:],
# d mu_a / dq_i dq_j
D[a][2] * (
scaling_factor[:, np.newaxis] *
scaling_factor[np.newaxis, :]
),
# d mu_a / dq_i dq_j dq_k
D[a][3] * (
scaling_factor[:, np.newaxis, np.newaxis] *
scaling_factor[np.newaxis, :, np.newaxis] *
scaling_factor[np.newaxis, np.newaxis, :]
)
]
for a in range(3) # loop over the x, y, and z axes
]
new_runner, _ = VPTRunner.construct(
runner.system.mol, # not actually used
runner.states.state_list,
potential_terms=v_expansion,
kinetic_terms=g_expansion,
pseudopotential_terms=u_expansion,
dipole_terms=d_expansion
)
runner.print_tables() # runs the code and prints the IR tables
new_runner.print_tables()
HOHPartialQuartic
def test_HOHPartialQuartic(self):
VPTRunner.helpers.run_anne_job(
# os.path.expanduser("~/Desktop/r_as"),
TestManager.test_data("vpt2_helpers_api/hod/x"),
# states=2, # max quanta to be focusing on
states=[[0, 0, 0], [0, 0, 1], [0, 1, 0]],
# max quanta to be focusing on
order=2, # None, # orderr of VPT
expansion_order=2, # None, # order of expansion of H can use {
# 'potential':int,
# 'kinetic':int,
# 'dipole':int}
# logger=filename
# mode_selection=[1, 2],
calculate_intensities=False,
zero_element_warning=False,
include_only_mode_couplings=[1, 2],
include_coriolis_coupling=False
# target_property='wavefunctions'
# return_runner=True
) # output file name
VPTRunner.helpers.run_anne_job(
# os.path.expanduser("~/Desktop/r_as"),
TestManager.test_data("vpt2_helpers_api/hod/x_no_bend"),
# states=2, # max quanta to be focusing on
states=[[0, 0, 0], [0, 0, 1], [0, 1, 0]],
# max quanta to be focusing on
order=2, # None, # orderr of VPT
expansion_order=2, # None, # order of expansion of H can use {
# 'potential':int,
# 'kinetic':int,
# 'dipole':int}
# logger=filename
# mode_selection=[1, 2],
calculate_intensities=False,
zero_element_warning=False,
# include_only_mode_couplings=[1, 2],
include_coriolis_coupling=True
# target_property='wavefunctions'
# return_runner=True
) # output file name
raise Exception(...)
VPTRunner.helpers.run_anne_job(
# os.path.expanduser("~/Desktop/r_as"),
TestManager.test_data("vpt2_helpers_api/hod/x_decoupled"),
# states=2, # max quanta to be focusing on
states=1,
# max quanta to be focusing on
order=2, # None, # orderr of VPT
expansion_order=2, # None, # order of expansion of H can use {
# 'potential':int,
# 'kinetic':int,
# 'dipole':int}
# logger=filename
# mode_selection=[1, 2],
calculate_intensities=False,
zero_element_warning=False,
include_coriolis_coupling=False
# target_property='wavefunctions'
# return_runner=True
) # output file name
"""
::> States Energies
> State Harmonic Anharmonic Harmonic Anharmonic
ZPE ZPE Frequency Frequency
0 0 0 4052.91097 4001.04707 - -
0 0 1 - - 3873.84521 3688.94993
0 1 0 - - 2810.03028 2723.42011
1 0 0 - - 1421.94645 1383.13454
"""
VPTRunner.helpers.run_anne_job(
# os.path.expanduser("~/Desktop/r_as"),
TestManager.test_data("vpt2_helpers_api/hod/x"),
# states=2, # max quanta to be focusing on
states=1,
# max quanta to be focusing on
order=2, # None, # orderr of VPT
expansion_order=2, # None, # order of expansion of H can use {
# 'potential':int,
# 'kinetic':int,
# 'dipole':int}
# logger=filename
mode_selection=[0, 2],
calculate_intensities=False,
zero_element_warning=False,
include_coriolis_coupling=False
# target_property='wavefunctions'
# return_runner=True
) # output file name
# runner3 = VPTRunner.helpers.run_anne_job(
# # os.path.expanduser("~/Desktop/r_as"),
# TestManager.test_data("vpt2_helpers_api/hod/x"),
# # states=2, # max quanta to be focusing on
# states=1,
# # max quanta to be focusing on
# order=2, # None, # orderr of VPT
# expansion_order=2, # None, # order of expansion of H can use {
# # 'potential':int,
# # 'kinetic':int,
# # 'dipole':int}
# # logger=filename
# # mode_selection=[1, 2],
# calculate_intensities=False,
# operator_coefficient_threshold=1e-12,
# zero_element_warning=False,
# # target_property='wavefunctions'
# return_runner=True
# ) # output file name
#
# runner4 = VPTRunner.helpers.run_anne_job(
# # os.path.expanduser("~/Desktop/r_as"),
# TestManager.test_data("vpt2_helpers_api/hod/x_sub"),
# # states=2, # max quanta to be focusing on
# states=1,
# # max quanta to be focusing on
# order=2, # None, # orderr of VPT
# expansion_order=2, # None, # order of expansion of H can use {
# # 'potential':int,
# # 'kinetic':int,
# # 'dipole':int}
# # logger=filename
# # mode_selection=[1, 2],
# calculate_intensities=False,
# operator_coefficient_threshold=-1,
# zero_element_warning=False
# # target_property='wavefunctions'
# , return_runner=True
# ) # output file name
"""
0 0 0 4052.91097 3994.84632 - -
0 0 1 - - 3873.84521 3685.79215
0 1 0 - - 2810.03028 2706.14630
1 0 0 - - 1421.94645 1383.40761
0 0 0 4052.91097 4033.89584 - -
0 0 1 - - 3873.84521 3697.97351
0 1 0 - - 2810.03028 2902.77195
1 0 0 - - 1421.94645 1380.70462
"""
# split1 = runner3[0].hamiltonian.get_Nielsen_energies([[0, 0, 0], [0, 0, 1]], return_split=True)
# split2 = runner4[0].hamiltonian.get_Nielsen_energies([[0, 0, 0], [0, 0, 1]], return_split=True)
# raise Exception(
# split1[2][0] - split2[2][0] # cubic contributions to X matrix
# )
# nie_full = np.sum(runner3[0].hamiltonian.get_Nielsen_energies([[0, 0, 0], [1, 0, 0], [0, 1, 0], [0, 0, 1]]), axis=0)
# nie_sub = np.sum(runner4[0].hamiltonian.get_Nielsen_energies([[0, 0, 0], [1, 0, 0], [0, 1, 0], [0, 0, 1]]), axis=0)
"""
array([1383.40761359, 2706.14629421, 3685.79215281])
array([1380.70462067, 2902.77194309, 3697.97351711]))
"""
raise Exception(...)
# (nie_full[1:] - nie_full[0]) * UnitsData.hartrees_to_wavenumbers,
# (nie_sub[1:] - nie_sub[0]) * UnitsData.hartrees_to_wavenumbers
# )
raise Exception(...)
"""
0 1973.85245 1992.46835 - -
1 - - 3947.70491 4042.24072
"""
runner1 = VPTRunner.helpers.run_anne_job(
os.path.expanduser("~/Desktop/r_as"),
# states=2, # max quanta to be focusing on
states=1,
# max quanta to be focusing on
order=2, # None, # orderr of VPT
expansion_order=2, # None, # order of expansion of H can use {
# 'potential':int,
# 'kinetic':int,
# 'dipole':int}
# logger=filename
calculate_intensities=False,
operator_coefficient_threshold=-1
# target_property='wavefunctions'
# return_runner=True
) # output file name
"""
State Frequency Intensity Frequency Intensity
0 0 1 3947.69802 75.46200 3761.26977 70.72838
0 1 0 3821.87392 5.56098 3652.31667 4.82837
1 0 0 1628.37574 0.00000 1590.97610 0.00483
State Frequency Intensity Frequency Intensity
0 0 1 3947.69802 75.46200 3874.95435 71.78714
0 1 0 3821.87392 0.00000 3864.33291 0.00111
1 0 0 1628.37574 0.00000 1620.53058 0.00003
"""
"""
::> States Energies
> State Harmonic Anharmonic Harmonic Anharmonic
ZPE ZPE Frequency Frequency
0 0 0 4698.97384 4628.17891 - -
0 0 1 - - 3947.69802 3761.26977
0 1 0 - - 3821.87392 3652.31667
1 0 0 - - 1628.37574 1590.97610
"""
runner2 = VPTRunner.helpers.run_anne_job(
os.path.expanduser("~/Desktop/r_a"),
# states=2, # max quanta to be focusing on
states=1,
# max quanta to be focusing on
order=2, # None, # orderr of VPT
expansion_order=2, # None, # order of expansion of H can use {
# 'potential':int,
# 'kinetic':int,
# 'dipole':int}
# logger=filename
calculate_intensities=False,
operator_coefficient_threshold=-1
# target_property='wavefunctions'
# return_runner=True
) # output file name
raise Exception(...)
raise Exception(
runner1[0].ham_opts.opts['potential_terms'][1],
runner2[0].ham_opts.opts['potential_terms'][1]
# runner2.ham_opts['potential_derivatives'],
)
HOHVPTNonGSRunner
def test_HOHVPTNonGSRunner(self):
file_name = "HOH_freq.fchk"
mol = Molecule.from_file(TestManager.test_data(file_name),
internals=[[0, -1, -1, -1], [1, 0, -1, -1], [2, 0, 1, -1]])
# raise Exception(mol.internal_coordinates)
ics = mol.internal_coordinates
derivs = mol.coords.jacobian(
ics.system,
[1, 2],
all_numerical=True,
converter_options={'reembed':True}
)
derivs = [x.reshape((9,)*(i+1) + (3, 3)) for i,x in enumerate(derivs)]
VPTRunner.run_simple(
TestManager.test_data(file_name),
2,
# target_property='wavefunctions',
# internals=mol.zmatrix,
initial_states=1,
operators={
'OH1':[ics[1, 0], derivs[0][:, 1, 0], derivs[1][:, :, 1, 0]],
'OH2':[ics[2, 0], derivs[0][:, 2, 0], derivs[1][:, :, 2, 0]],
'HOH':[ics[2, 1], derivs[0][:, 2, 1], derivs[1][:, :, 2, 1]]
},
logger=True
)
HOHVPTRunnerFlow
def test_HOHVPTRunnerFlow(self):
file_name = "HOH_freq.fchk"
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
memory_constrained=True,
logger=True
)
system = VPTSystem(TestManager.test_data(file_name))
states = VPTStateSpace.from_system_and_quanta(system, 3)
pt_opts = VPTSolverOptions(state_space_filters=states.get_filter("intensities"))
run_opts = VPTRuntimeOptions(logger=True)
runner = VPTRunner(system, states, runtime_options=run_opts, solver_options=pt_opts)
runner.print_tables()
HOHVPTRunnerShifted
def test_HOHVPTRunnerShifted(self):
file_name = "HOH_freq.fchk"
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
logger=True,
degeneracy_specs='auto',
corrected_fundamental_frequencies=np.array([1600, 3775, 3880])/UnitsData.convert("Hartrees", "Wavenumbers")
)
OCHHVPTRunnerShifted
def test_OCHHVPTRunnerShifted(self):
file_name = "OCHH_freq.fchk"
VPTRunner.run_simple(
TestManager.test_data(file_name),
2,
logger=True,
degeneracy_specs='auto',
corrected_fundamental_frequencies=np.array([1188, 1252, 1527, 1727, 2977, 3070]) / UnitsData.convert("Hartrees", "Wavenumbers")
)
HOONOVPTRunnerShifted
def test_HOONOVPTRunnerShifted(self):
file_name = "HOONO_freq.fchk"
VPTRunner.run_simple(
TestManager.test_data(file_name),
2,
logger=True,
degeneracy_specs='auto',
corrected_fundamental_frequencies=np.array([
355.73348, 397.16760, 524.09935,
715.88331, 836.39478, 970.87676,
1433.60940, 1568.50215, 3486.85528
]) / UnitsData.convert("Hartrees", "Wavenumbers")
)
CrieegeeVPTRunnerShifted
def test_CrieegeeVPTRunnerShifted(self):
# with BlockProfiler('Crieegee', print_res=True):
freqs = VPTSystem('criegee_eq_anh.fchk').mol.normal_modes.modes.freqs
freqs = freqs.copy()
freqs[1] += 10/UnitsData.convert("Hartrees", "Wavenumbers")
VPTRunner.run_simple(
# 'criegee_eq_anh.fchk',
2,
logger=True,
degeneracy_specs='auto',
corrected_fundamental_frequencies=freqs
# corrected_fundamental_frequencies=np.array([
# 200.246, 301.985 + 10, 462.536, 684.792, 736.234, 961.474, 984.773, 1038.825, 1120.260, 1327.450, 1402.397,
# 1449.820, 1472.576, 1519.875, 3037.286, 3078.370, 3174.043, 3222.828
# ])/UnitsData.convert("Hartrees", "Wavenumbers")
)
HOHVPTRunner3rd
def test_HOHVPTRunner3rd(self):
"""
test that runner works for 3rd order PT, too
:return:
:rtype:
"""
file_name = "HOH_freq.fchk"
handling_mode="unhandled"
logger=Logger()
with logger.block(tag="Internals 2nd Order triad"):
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
logger=logger,
order=2,
internals=[
[0, -1, -1, -1],
[1, 0, -1, -1],
[2, 0, 1, -1]
],
expansion_order=2,
degeneracy_specs=[
[[0, 0, 1], [2, 0, 0]],
[[0, 1, 0], [2, 0, 0]],
]
)
with logger.block(tag="Internals 3rd Order triad"):
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
logger=logger,
order=3,
internals=[
[0, -1, -1, -1],
[1, 0, -1, -1],
[2, 0, 1, -1]
],
expansion_order=2,
degeneracy_specs=[
[[0, 0, 1], [2, 0, 0]],
[[0, 1, 0], [2, 0, 0]],
]
)
with logger.block(tag="Internals 2nd Order dyad"):
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
logger=logger,
order=2,
internals=[
[0, -1, -1, -1],
[1, 0, -1, -1],
[2, 0, 1, -1]
],
expansion_order=2,
degeneracy_specs=[
[[0, 1, 0], [2, 0, 0]],
]
)
with logger.block(tag="Internals 3rd Order dyad"):
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
logger=logger,
order=3,
internals=[
[0, -1, -1, -1],
[1, 0, -1, -1],
[2, 0, 1, -1]
],
expansion_order=2,
degeneracy_specs=[
[[0, 1, 0], [2, 0, 0]],
]
)
with logger.block(tag="Cartesians 2nd Order triad"):
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
logger=logger,
order=2,
expansion_order=2,
degeneracy_specs=[
[[0, 0, 1], [2, 0, 0]],
[[0, 1, 0], [2, 0, 0]],
]
)
with logger.block(tag="Cartesians 3rd Order triad"):
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
logger=logger,
order=3,
expansion_order=2,
mixed_derivative_handling_mode=handling_mode,
degeneracy_specs=[
[[0, 0, 1], [2, 0, 0]],
[[0, 1, 0], [2, 0, 0]],
]
)
with logger.block(tag="Cartesians 2nd Order dyad"):
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
logger=logger,
order=2,
expansion_order=2,
degeneracy_specs=[
[[0, 1, 0], [2, 0, 0]],
]
)
with logger.block(tag="Cartesians 3rd Order dyad"):
VPTRunner.run_simple(
TestManager.test_data(file_name),
3,
logger=logger,
order=3,
expansion_order=2,
degeneracy_specs=[
[[0, 1, 0], [2, 0, 0]],
]
)
GetDegenerateSpaces
def test_GetDegenerateSpaces(self):
base_states = [
[0, 0, 1],
[0, 1, 0],
[0, 2, 1],
[0, 4, 0]
]
degenerate_states = VPTStateSpace.get_degenerate_polyad_space(
base_states,
[
[
[0, 2, 0],
[0, 0, 1]
]
],
)
ClHOClRunner
def test_ClHOClRunner(self):
file_name = "cl_hocl.fchk"
state = VPTStateMaker(6)
COM = -3
A = -2
C = -1
_ = 1000
O = 0
H = 1
Cl = 2
X = 3
VPTRunner.run_simple(
TestManager.test_data(file_name),
[
state(),
state([1, 1]),
state([1, 2]),
state([1, 3]),
state([1, 2], [5, 1]),
state([1, 1], [2, 2]),
],
logger=True,
handle_strong_couplings=True,
strong_coupling_test_modes=list(range(3, 6))
)
"""
> [[0 0 0 0 0 2]
> [1 0 0 0 0 2]
> [0 1 0 0 0 2]
> [0 0 0 0 2 1]
> [0 0 0 0 4 0]
> [0 2 0 0 0 2]
> [0 1 0 0 2 1]]
Harmonic Anharmonic
State Frequency Intensity Frequency Intensity
0 0 0 0 0 1 2709.16096 2782.25433 2285.38768 2611.96281
0 0 0 0 0 2 5418.32192 0.00000 4140.45935 19.13726
0 0 0 0 0 3 8127.48288 0.00000 5353.97008 0.16004
0 1 0 0 0 2 5699.88024 0.00000 4605.01290 3.93389
0 0 0 0 2 1 5592.48466 0.00000 5023.09956 7.99053
"""
VPTRunner.run_simple(
TestManager.test_data(file_name),
[
state(),
state([1, 1]),
state([1, 2]),
state([1, 3]),
state([1, 2], [5, 1]),
state([1, 1], [2, 2]),
],
logger=True,
handle_strong_couplings=True
)
"""
> [[0 0 0 0 0 2]
> [0 0 0 0 2 1]
> [0 0 0 0 4 0]]
Harmonic Anharmonic
State Frequency Intensity Frequency Intensity
0 0 0 0 0 1 2709.16096 2782.25433 2285.38768 2611.96281
0 0 0 0 0 2 5418.32192 0.00000 4096.10015 21.49976
0 0 0 0 0 3 8127.48288 0.00000 5353.97008 0.16004
0 1 0 0 0 2 5699.88024 0.00000 4374.63719 2.66632
0 0 0 0 2 1 5592.48466 0.00000 4964.16010 6.86648
"""
VPTRunner.run_simple(
TestManager.test_data(file_name),
[
state(),
state([1, 1]),
state([1, 2]),
state([1, 3]),
state([1, 2], [5, 1]),
state([1, 1], [2, 2]),
],
logger=True,
handle_strong_couplings=True
, strong_coupling_test_modes=list(range(3, 6))
, internals=[
[Cl, _, _, _],
[O, Cl, _, _],
[X, O, Cl, _],
[H, O, Cl, X],
]
)
"""
> [[0 0 0 0 0 2]
> [1 0 0 0 0 2]
> [0 1 0 0 0 2]
> [0 0 0 0 2 1]
> [0 0 0 0 4 0]
> [0 2 0 0 0 2]
> [0 1 0 0 2 1]]
Harmonic Anharmonic
State Frequency Intensity Frequency Intensity
0 0 0 0 0 1 2709.16096 2782.25433 2305.91708 2613.72238
0 0 0 0 0 2 5418.32192 0.00000 4174.53466 17.82679
0 0 0 0 0 3 8127.48288 0.00000 5353.97246 0.16004
0 1 0 0 0 2 5699.88024 0.00000 4645.21985 6.56429
0 0 0 0 2 1 5592.48466 0.00000 5114.27886 9.19226
"""
VPTRunner.run_simple(
TestManager.test_data(file_name),
[
state(),
state([1, 1]),
state([1, 2]),
state([1, 3]),
state([1, 2], [5, 1]),
state([1, 1], [2, 2]),
],
logger=True,
handle_strong_couplings=True
, internals=[
[Cl, _, _, _],
[O, Cl, _, _],
[X, O, Cl, _],
[H, O, Cl, X],
]
)
"""
> [[0 0 0 0 0 2]
> [0 0 0 0 2 1]
> [0 0 0 0 4 0]]
Harmonic Anharmonic
State Frequency Intensity Frequency Intensity
0 0 0 0 0 1 2709.16096 2782.25433 2305.91708 2613.72238
0 0 0 0 0 2 5418.32192 0.00000 4130.15930 22.20869
0 0 0 0 0 3 8127.48288 0.00000 5353.97246 0.16004
0 1 0 0 0 2 5699.88024 0.00000 4374.63638 2.66634
0 0 0 0 2 1 5592.48466 0.00000 5053.08138 7.79496
"""
AnalyticModels
def test_AnalyticModels(self):
from Psience.AnalyticModels import AnalyticModel as Model
from McUtils.Data import AtomData, UnitsData
order = 4
# include_potential=False
# include_gmatrix=True
# include_pseudopotential=True
expansion_order={
'potential':0,
'gmatrix':4,
'pseudopotential':4
}
hoh_params = {}
hoh_params["mH"] = AtomData["H"]["Mass"] * UnitsData.convert("AtomicMassUnits", "AtomicUnitOfMass")
hoh_params["mO"] = AtomData["O"]["Mass"] * UnitsData.convert("AtomicMassUnits", "AtomicUnitOfMass")
# morse stretch parameters
cm2borh = UnitsData.convert("Angstroms", "BohrRadius")
hoh_params['re'] = 0.9575 * cm2borh
erg2h = UnitsData.convert("Ergs", "Hartrees")
invcm2borh = UnitsData.convert("InverseAngstroms", "InverseBohrRadius")
hoh_params['De'] = 8.84e-12 * erg2h
hoh_params['a'] = 2.175 * invcm2borh
# harmonic bend parameters
hoh_params['b_e'] = np.deg2rad(104.5)
hoh_params['k_b'] = 3.2 ** 2 * 1600 * UnitsData.convert("Wavenumbers",
"Hartrees") # the 3.2 is some deep magic I don't understand
model = Model(
[
Model.r(1, 2),
Model.r(2, 3),
Model.a(1, 2, 3)
],
Model.Potential.morse(1, 2,
De=hoh_params["De"],
a=hoh_params["a"],
re=hoh_params["re"]
)
+ Model.Potential.morse(2, 3,
De=hoh_params["De"],
a=hoh_params["a"],
re=hoh_params["re"]
)
+ Model.Potential.harmonic(1, 2, 3,
k=hoh_params["k_b"],
qe=hoh_params["b_e"]
),
# dipole=Model.r(1, 2) + Model.r(2, 3),
values={
Model.m(1): hoh_params["mH"],
Model.m(2): hoh_params["mO"],
Model.m(3): hoh_params["mH"],
Model.r(1, 2): hoh_params["re"],
Model.r(2, 3): hoh_params["re"],
Model.a(1, 2, 3): hoh_params["b_e"]
}
)
# raise Exception(model.g())
model.run_VPT(order=order, return_analyzer=False, expansion_order=expansion_order)
class harmonically_coupled_morse:
# mass_weights = masses[:2] / np.sum(masses[:2])
def __init__(self,
De_1, a_1, re_1,
De_2, a_2, re_2,
kb, b_e
):
self.De_1 = De_1
self.a_1 = a_1
self.re_1 = re_1
self.De_2 = De_2
self.a_2 = a_2
self.re_2 = re_2
self.kb = kb
self.b_e = b_e
def __call__(self, carts):
v1 = carts[..., 1, :] - carts[..., 0, :]
v2 = carts[..., 2, :] - carts[..., 0, :]
r1 = nput.vec_norms(v1) - self.re_1
r2 = nput.vec_norms(v2) - self.re_2
bend, _ = nput.vec_angles(v1, v2)
bend = bend - self.b_e
return (
self.De_1 * (1 - np.exp(-self.a_1 * r1)) ** 2
+ self.De_2 * (1 - np.exp(-self.a_2 * r2)) ** 2
+ self.kb * bend ** 2
)
atoms = ["O", "H", "H"]
coords = np.array([
[0.000000, 0.000000, 0.000000],
[hoh_params["re"], 0.000000, 0.000000],
np.dot(
nput.rotation_matrix([0, 0, 1], hoh_params["b_e"]),
[hoh_params["re"], 0.000000, 0.000000]
)
])
masses = np.array([AtomData[x]["Mass"] for x in atoms]) * UnitsData.convert("AtomicMassUnits", "AtomicUnitOfMass")
pot_file = os.path.expanduser('~/Desktop/water_pot.hdf5')
water_chk = Checkpointer.from_file(pot_file)
if expansion_order['potential'] > -1:
with water_chk as wat:
try:
potential_derivatives = wat['potential_derivatives']
except (OSError, KeyError):
potential_function = harmonically_coupled_morse(
hoh_params["De"], hoh_params["a"], hoh_params["re"],
hoh_params["De"], hoh_params["a"], hoh_params["re"],
hoh_params["k_b"], hoh_params["b_e"]
)
deriv_gen = FiniteDifferenceDerivative(potential_function,
function_shape=((None, None), 0),
stencil=5 + expansion_order['potential'],
mesh_spacing=1e-3,
).derivatives(coords)
potential_derivatives = deriv_gen.derivative_tensor(list(range(1, order + 3)))
wat['potential_derivatives'] = potential_derivatives
else:
potential_derivatives = []
# analyzer = VPTRunner.run_simple(
# [atoms, coords, dict(masses=masses)],
# 2,
# potential_derivatives=potential_derivatives,
# calculate_intensities=False,
# order=order,
# include_potential=include_potential,
# include_gmatrix=include_gmatrix,
# include_pseudopotential=include_pseudopotential,
# include_coriolis_coupling=include_gmatrix
# )
# analyzer.print_output_tables(print_intensities=False, print_energies=True)
# try:
# os.remove(os.path.expanduser('~/Desktop/water_analyt.hdf5'))
# except:
# pass
analyzer = VPTRunner.run_simple(
[atoms, coords, dict(masses=masses)],
2,
potential_derivatives=potential_derivatives,
calculate_intensities=False,
internals=[
[0, -1, -1, -1],
[1, 0, -1, -1],
[2, 0, 1, -1]
],
order=order,
internal_fd_mesh_spacing=1e-2,
cartesian_fd_mesh_spacing=1e-2,
checkpoint=os.path.expanduser('~/Desktop/water_analyt.hdf5'),
expansion_order=expansion_order
)
HOHCorrectedDegeneracies
def test_HOHCorrectedDegeneracies(self):
VPTRunner.run_simple(
TestManager.test_data('HOH_freq.fchk'),
2,
zero_order_energy_corrections=[
[(0, 1, 0), (4681.56364+3800) * UnitsData.convert("Wavenumbers", "Hartrees")],
[(0, 2, 0), (4681.56364+7800) * UnitsData.convert("Wavenumbers", "Hartrees")],
[(0, 0, 2), (4681.56364+7801) * UnitsData.convert("Wavenumbers", "Hartrees")],
[(0, 3, 1), (4681.56364+3801) * UnitsData.convert("Wavenumbers", "Hartrees")],
],
degeneracy_specs={
'wfc_threshold':.3,
'extra_groups':[
[[0, 2, 0], [0, 1, 1]],
[[0, 0, 2], [0, 3, 1]],
]
}
# operator_chunk_size=int(12)
)
HOHCorrectedPostfilters
def test_HOHCorrectedPostfilters(self):
# VPTAnalyzer.run_VPT(
# TestManager.test_data('HOH_freq.fchk'),
# 2,
# zero_order_energy_corrections=[
# [(0, 1, 0), (4681.56364 + 3800) * UnitsData.convert("Wavenumbers", "Hartrees")],
# [(0, 2, 0), (4681.56364 + 7800) * UnitsData.convert("Wavenumbers", "Hartrees")],
# [(0, 0, 2), (4681.56364 + 7801) * UnitsData.convert("Wavenumbers", "Hartrees")],
# ],
# degeneracy_specs='auto',
# basis_filters={'max_quanta':[0, -1, -1]}
# ).print_output_tables()
VPTAnalyzer.run_VPT(
TestManager.test_data('HOH_freq.fchk'),
2,
zero_order_energy_corrections=[
[(0, 1, 0), (4681.56364 + 3800) * UnitsData.convert("Wavenumbers", "Hartrees")],
[(0, 2, 0), (4681.56364 + 7800) * UnitsData.convert("Wavenumbers", "Hartrees")],
[(0, 0, 2), (4681.56364 + 7801) * UnitsData.convert("Wavenumbers", "Hartrees")],
],
degeneracy_specs='auto',
basis_filters=[
{'max_quanta': [2, -1, -1]},
{'excluded_transitions': [[1, 0, 0], [0, 1, 0], [0, 0, 1]]}
]
).print_output_tables()
WaterSkippedCouplings
def test_WaterSkippedCouplings(self):
VPTRunner.run_simple(
TestManager.test_data('water_freq.fchk'),
1,
degeneracy_specs='auto',
operator_coefficient_threshold=(1.0e-8)
)
H2COPolyads
def test_H2COPolyads(self):
internals = [
[0, -1, -1, -1],
[1, 0, -1, -1],
[2, 1, 0, -1],
[3, 1, 0, 2]
]
VPTRunner.run_simple(
TestManager.test_data('OCHH_freq.fchk'),
2,
degeneracy_specs=True
# degeneracy_specs={
# 'nT':[1, 1, 1, 1, 2, 2]
# }
)
H2COModeSel
def test_H2COModeSel(self):
internals = [
[0, -1, -1, -1],
[1, 0, -1, -1],
[2, 1, 0, -1],
[3, 1, 0, 2]
]
VPTRunner.run_simple(
TestManager.test_data('OCHH_freq.fchk'),
1,
degeneracy_specs=None,
mode_selection=[1, 2, 3, 4, 5]
)
HODRephase
def test_HODRephase(self):
VPTRunner.run_simple(
TestManager.test_data('HOD_freq_16.fchk'),
1,
degeneracy_specs=None,
# order=4,
# expansion_order=2
)
HOHRephase
def test_HOHRephase(self):
VPTRunner.run_simple(
TestManager.test_data('HOH_freq.fchk'),
1,
degeneracy_specs=None,
# order=4,
# expansion_order=2
)
NH3
def test_NH3(self):
VPTRunner.run_simple(
TestManager.test_data('nh3.fchk'),
2,
# degeneracy_specs=False,
order=4,
expansion_order=2,
# basis_filters={
# 'max_quanta':[2, -1, -1, -1, -1, -1]
# }
)
HOONO
def test_HOONO(self):
VPTRunner.run_simple(
TestManager.test_data('HOONO_freq.fchk'),
1,
degeneracy_specs=None,
# order=4,
# expansion_order=2
)
H2COSkippedCouplings
def test_H2COSkippedCouplings(self):
VPTRunner.run_simple(
TestManager.test_data('OCHH_freq.fchk'),
1,
degeneracy_specs='auto',
operator_coefficient_threshold=1.00 / 219475
)
WaterDimerSkippedCouplings
def test_WaterDimerSkippedCouplings(self):
COM = -3
A = -2
C = -1
X = 1000
LHF = 0
LO = 1
SH = 2
RO = 3
RH1 = 4
RH2 = 5
internals = [
[LHF, X, X, X],
[LO, LHF, X, X],
[SH, LO, LHF, X],
[RH2, SH, LO, LHF], # get out of plane
[RO, LO, RH2, LHF],
[RH1, RO, RH2, LHF]
]
VPTRunner.run_simple(
TestManager.test_data('water_dimer_freq.fchk'),
1,
degeneracy_specs='auto',
# operator_coefficient_threshold=0.005/219475,
# basis_filters=[
# {'max_quanta': [3, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1, -1]}
# ],
# internals=internals,
mode_selection=[1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11]
)
OCHHInternals
def test_OCHHInternals(self):
tag = 'OCHH Internals'
file_name = "OCHH_freq.fchk"
zmatrix = [
[0, -1, -1, -1],
[1, 0, -1, -1],
[2, 1, 0, -1],
[3, 1, 0, 2]
]
def conv(r, t, f, **kwargs):
return np.array([r**2+1, t, f])
def inv(r2, t, f, **kwargs):
return np.array([np.sqrt(r2-1), t, f])
# def conv(crds, **kwargs):
# return crds
# def inv(crds, **kwargs):
# return crds
internals = {
'zmatrix':zmatrix,
'conversion':conv,
'inverse':inv,
# 'converter_options':{
# 'pointwise':False,
# # 'jacobian_prep':ZMatrixCoordinateSystem.jacobian_prep_coordinates
# }
}
print("Cart:")
# VPTAnalyzer.run_VPT(TestManager.test_data(file_name), 2).print_output_tables()
print("ZMat:")
# VPTAnalyzer.run_VPT(TestManager.test_data(file_name), 2, internals=zmatrix).print_output_tables()
print("Custom:")
# VPTRunner.run_simple(TestManager.test_data(file_name), 2, internals=internals)
VPTAnalyzer.run_VPT(TestManager.test_data(file_name), 2, internals=internals).print_output_tables()
NH3Units
def test_NH3Units(self):
tag = 'NH3 Internals'
file_name = "nh3.fchk"
_ = -1
zmatrix = [
[0, _, _, _],
[1, 0, _, _],
[2, 0, 1, _],
[3, 0, 1, 2]
]
def conv(r, t, f, **kwargs):
cp1 = np.cos(f[..., 3]) # skip three for embedding
ct1 = np.cos(t[..., 2]) # skip two for embedding
ct2 = np.cos(t[..., 3])
st1 = np.sin(t[..., 2])
st2 = np.sin(t[..., 3])
f[..., 3] = np.arccos(st1*st2*cp1+ct1*ct2)
return np.array([r, t, f])
def inv(r, t, f, **kwargs):
cp1 = np.cos(f[..., 3])
ct1 = np.cos(t[..., 2])
ct2 = np.cos(t[..., 3])
st1 = np.sin(t[..., 2])
st2 = np.sin(t[..., 3])
f[..., 3] = np.arccos((cp1-ct1*ct2)/(st1*st2))
return np.array([r, t, f])
# def conv(crds, **kwargs):
# return crds
# def inv(crds, **kwargs):
# return crds
# internals = {
# 'zmatrix':zmatrix,
# 'conversion':conv,
# 'inverse':inv,
# # 'converter_options':{
# # 'pointwise':False,
# # # 'jacobian_prep':ZMatrixCoordinateSystem.jacobian_prep_coordinates
# # }
# }
file = TestManager.test_data(file_name)
print("Cart:")
VPTAnalyzer.run_VPT(file, 2).print_output_tables()
print("ZMat:")
# VPTAnalyzer.run_VPT(TestManager.test_data(file_name), 2, internals=zmatrix).print_output_tables()
print("Custom:")
# VPTRunner.run_simple(TestManager.test_data(file_name), 2, internals=internals)
# VPTAnalyzer.run_VPT(TestManager.test_data(file_name), 2, internals=internals, handle_strong_couplings=True).print_output_tables()
""" With Cartesian coordinates
Harmonic Anharmonic
State Frequency Intensity Frequency Intensity
0 0 0 0 0 1 3649.84753 8.40063 3673.97101 8.16596
0 0 0 0 1 0 3649.84749 8.40063 3673.99153 8.16597
0 0 0 1 0 0 3502.88652 3.39049 3504.89231 4.29459
0 0 1 0 0 0 1668.90366 14.31874 1611.87387 15.15334
0 1 0 0 0 0 1668.90366 14.31874 1611.87470 15.15358
1 0 0 0 0 0 1037.51781 139.18086 907.20372 146.77249
0 0 0 0 0 2 7299.69506 0.00000 7358.46413 0.23938
0 0 0 0 2 0 7299.69499 0.00000 7322.01149 0.00313
0 0 0 2 0 0 7005.77304 0.00000 6948.64326 0.01480
0 0 2 0 0 0 3337.80732 0.00000 3216.64730 0.29740
0 2 0 0 0 0 3337.80733 0.00000 3191.38576 0.04236
2 0 0 0 0 0 2075.03561 0.00000 1716.91900 0.05990
0 0 0 0 1 1 7299.69502 0.00000 7541.05092 0.25778
0 0 0 1 0 1 7152.73405 0.00000 7166.06224 0.21966
0 0 1 0 0 1 5318.75119 0.00000 5240.10874 0.00432
0 1 0 0 0 1 5318.75119 0.00000 5279.00970 1.21013
1 0 0 0 0 1 4687.36534 0.00000 4547.87059 0.68588
0 0 0 1 1 0 7152.73402 0.00000 7202.90789 0.20904
0 0 1 0 1 0 5318.75115 0.00000 5274.40898 0.00033
0 1 0 0 1 0 5318.75116 0.00000 5235.65311 1.20013
1 0 0 0 1 0 4687.36530 0.00000 4547.88276 0.68588
0 0 1 1 0 0 5171.79018 0.00000 5099.83410 0.04409
0 1 0 1 0 0 5171.79019 0.00000 5099.83542 0.04422
1 0 0 1 0 0 4540.40433 0.00000 4425.74209 0.84746
0 1 1 0 0 0 3337.80732 0.00000 3216.43526 0.29740
1 0 1 0 0 0 2706.42147 0.00000 2512.73682 0.00177
1 1 0 0 0 0 2706.42147 0.00000 2512.73850 0.00177
0 2 0 1 0 0 6840.69385 0.00000 6713.79845 0.00011
0 0 2 1 0 0 6840.69384 0.00000 6698.61681 0.00040
0 3 0 0 0 0 5006.71099 0.00000 4789.39177 0.00714
0 0 3 0 0 0 5006.71098 0.00000 4789.38260 0.00626
"""
""" With regular Z-matrix coordinates
============================================= IR Data ==============================================
Harmonic Anharmonic
State Frequency Intensity Frequency Intensity
0 0 0 0 0 1 3649.84753 8.40063 3726.14107 8.01000
0 0 0 0 1 0 3649.84749 8.40063 3720.43203 8.14962
0 0 0 1 0 0 3502.88652 3.39049 3504.98059 4.27590
0 0 1 0 0 0 1668.90366 14.31874 1635.12777 15.11316
0 1 0 0 0 0 1668.90367 14.31874 1634.58245 15.13005
1 0 0 0 0 0 1037.51781 139.18086 951.20470 148.04683
0 0 0 0 0 2 7299.69506 0.00000 7443.94606 0.24253
0 0 0 0 2 0 7299.69499 0.00000 7420.05073 0.00705
0 0 0 2 0 0 7005.77304 0.00000 6958.79881 0.01981
0 0 2 0 0 0 3337.80732 0.00000 3263.93164 0.30176
0 2 0 0 0 0 3337.80733 0.00000 3234.19765 0.04307
2 0 0 0 0 0 2075.03562 0.00000 1804.93029 0.06297
0 0 0 0 1 1 7299.69502 0.00000 7636.09541 0.26285
0 0 0 1 0 1 7152.73405 0.00000 7227.52952 0.22936
0 0 1 0 0 1 5318.75119 0.00000 5338.82560 0.28104
0 1 0 0 0 1 5318.75120 0.00000 5378.12318 1.09984
1 0 0 0 0 1 4687.36534 0.00000 4697.63552 0.70847
0 0 0 1 1 0 7152.73402 0.00000 7257.29782 0.19690
0 0 1 0 1 0 5318.75115 0.00000 5374.55183 0.13318
0 1 0 0 1 0 5318.75116 0.00000 5333.95854 0.94634
1 0 0 0 1 0 4687.36530 0.00000 4675.38562 0.70511
0 0 1 1 0 0 5171.79018 0.00000 5128.93044 0.04608
0 1 0 1 0 0 5171.79019 0.00000 5129.48827 0.04659
1 0 0 1 0 0 4540.40433 0.00000 4469.82653 0.85590
0 1 1 0 0 0 3337.80732 0.00000 3257.94962 0.30112
1 0 1 0 0 0 2706.42147 0.00000 2577.72518 0.00182
1 1 0 0 0 0 2706.42147 0.00000 2579.04652 0.00182
0 3 0 0 0 0 5006.71100 0.00000 4848.28115 0.00859
0 0 3 0 0 0 5006.71098 0.00000 4848.52584 0.00458
"""
""" With symmetrized coordinates
State Frequency Intensity Frequency Intensity
0 0 0 0 0 1 3649.84753 8.40063 3723.70074 8.02349
0 0 0 0 1 0 3649.84749 8.40063 3723.72163 8.02350
0 0 0 1 0 0 3502.88652 3.39049 3504.97874 4.27470
0 0 1 0 0 0 1668.90366 14.31874 1634.65206 15.13159
0 1 0 0 0 0 1668.90367 14.31874 1634.64634 15.13156
1 0 0 0 0 0 1037.51781 139.18086 952.40922 148.07587
0 0 0 0 0 2 7299.69506 0.00000 7444.16390 0.24658
0 0 0 0 2 0 7299.69499 0.00000 7421.28673 0.00317
0 0 0 2 0 0 7005.77305 0.00000 6958.79513 0.01981
0 0 2 0 0 0 3337.80732 0.00000 3263.57753 0.30173
0 2 0 0 0 0 3337.80733 0.00000 3233.82922 0.04292
2 0 0 0 0 0 2075.03561 0.00000 1807.33816 0.06305
0 0 0 0 1 1 7299.69503 0.00000 7637.00847 0.26180
0 0 0 1 0 1 7152.73405 0.00000 7229.66109 0.21733
0 0 1 0 0 1 5318.75119 0.00000 5339.24402 0.00449
0 1 0 0 0 1 5318.75120 0.00000 5376.53040 1.23248
1 0 0 0 0 1 4687.36534 0.00000 4689.39755 0.70723
0 0 0 1 1 0 7152.73402 0.00000 7256.19539 0.20990
0 0 1 0 1 0 5318.75115 0.00000 5373.43275 0.00026
0 1 0 0 1 0 5318.75116 0.00000 5336.29143 1.22319
1 0 0 0 1 0 4687.36530 0.00000 4689.40994 0.70723
0 0 1 1 0 0 5171.79018 0.00000 5129.06995 0.04504
0 1 0 1 0 0 5171.79019 0.00000 5129.06553 0.04502
1 0 0 1 0 0 4540.40433 0.00000 4471.02511 0.85613
0 1 1 0 0 0 3337.80733 0.00000 3257.49832 0.30121
1 0 1 0 0 0 2706.42147 0.00000 2579.33175 0.00182
1 1 0 0 0 0 2706.42147 0.00000 2579.32464 0.00182
0 3 0 0 0 0 5006.71100 0.00000 4847.85818 0.00744
0 0 3 0 0 0 5006.71098 0.00000 4847.85797 0.00578
"""
AnneAPI
def test_AnneAPI(self):
VPTRunner.run_simple(
TestManager.test_data('HOD_freq_16.fchk'), 2,
# calculate_intensities=False
)
# raise Exception("...")
# test_folder = TestManager.test_data('vpt2_helpers_api/hod')
# runner, dat = VPTRunner.construct(TestManager.test_data('HOD_freq_16.fchk'), 2)
# fuck = dat[0]
# raise Exception(fuck.mol.normal_modes.modes.basis.matrix.T)
# fuck = runner.hamiltonian
# shit_strings = []
# cub = fuck.V_terms[1]
# h2w = UnitsData.convert("Hartrees", "Wavenumbers")
# for i in range(cub.shape[0]):
# for j in range(i, cub.shape[1]):
# for k in range(j, cub.shape[2]):
# shit_strings.append(f"{i+1} {j+1} {k+1} {cub[i, j ,k]*h2w}")
# fuck = runner.hamiltonian
# shit_strings = []
# quart = fuck.V_terms[2]
# h2w = UnitsData.convert("Hartrees", "Wavenumbers")
# for i in range(quart.shape[0]):
# for j in range(i, quart.shape[2]):
# for k in range(j, quart.shape[2]):
# for l in range(k, quart.shape[3]):
# shit_strings.append(f"{i+1} {j+1} {k+1} {l+1} {quart[i, j, k, l]*h2w}")
# #
# raise Exception("\n".join(shit_strings))
# fuck = runner.get_wavefunctions()
# shit_strings = []
# dts = fuck.dipole_terms
#
# lints = np.array([d[1] for d in dts])
# for a in range(lints.shape[0]):
# for i in range(lints.shape[1]):
# shit_strings.append(f"{a+1} {i+1} {lints[a, i]}")
# shit_strings.append("")
#
# dints = np.array([d[2] for d in dts])
# for a in range(dints.shape[0]):
# for i in range(dints.shape[1]):
# for j in range(i, dints.shape[2]):
# shit_strings.append(f"{a + 1} {i + 1} {j + 1} {dints[a, i, j]}")
# shit_strings.append("")
#
# cints = np.array([d[3] for d in dts])
# for a in range(cints.shape[0]):
# for i in range(cints.shape[1]):
# for j in range(i, cints.shape[2]):
# for k in range(j, cints.shape[3]):
# shit_strings.append(f"{a + 1} {i + 1} {j + 1} {k + 1} {cints[a, i, j, k]}")
# #
# raise Exception("\n".join(shit_strings))
VPTRunner.helpers.run_anne_job(
TestManager.test_data('vpt2_helpers_api/hod/x'),
# calculate_intensities=False,
# expansion_order=2
)